Georeactor Blog
RSS FeedQuinoa news + genome browser
When writing the closing for my quinoa presentation at CSV Conf, I was thinking about a productive way to continue or conclude my project. Eventually I took a deeper look at public genome browsers - a way to navigate the genome and visualize the locations of specific annotated genes.
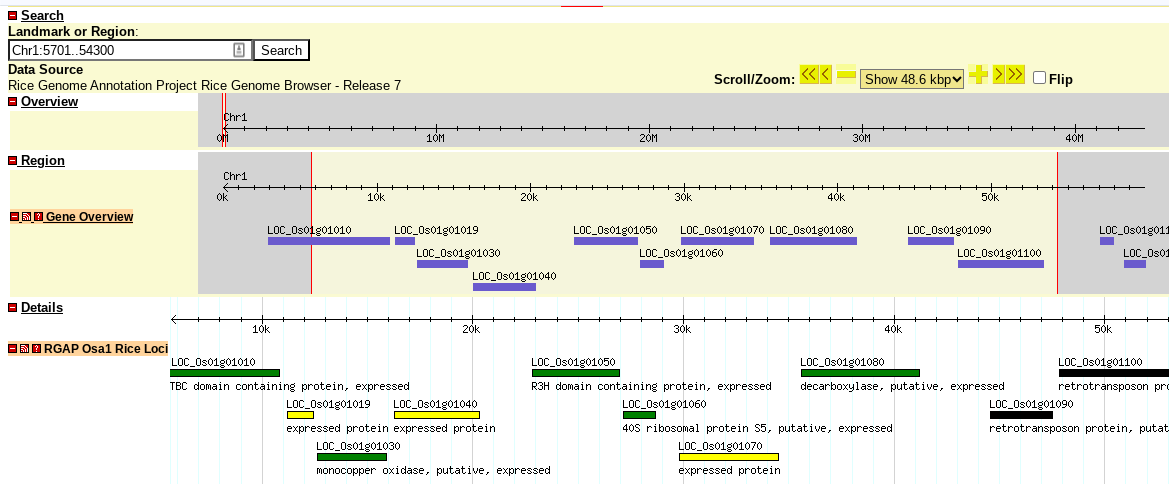
Major crops like rice and maize have websites dedicated to this (above is https://rice.uga.edu ), and more obscure crops like quinoa don't. The larger context is unclear - maybe this is an older way of doing things, or they are being run internally, or maybe the community lacks resources to host a public site. What I can do is:
- download quinoa genome
- find an open source genome browser to display the genome
- add my own annotations / model predictions
- attempt to build a pipeline to take in research papers and output annotations
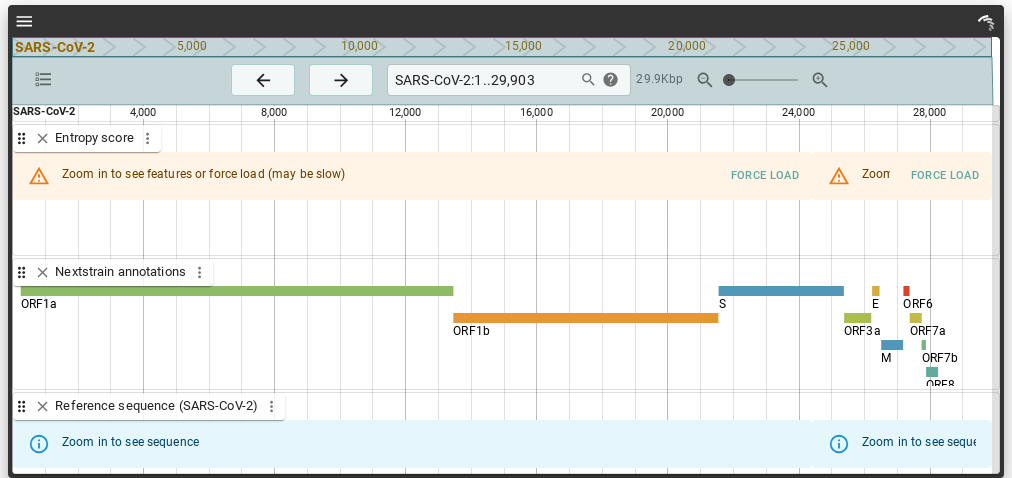
The go-to open source tool for this is jBrowse2, built on Yarn / React / SVG. This is code which I understand and could maybe contribute to. Here's a look:
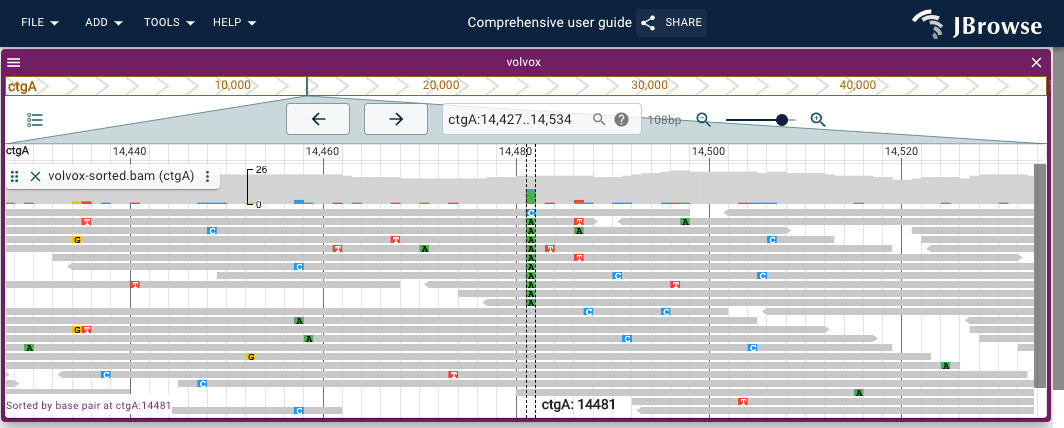
jBrowse has other views and features which I don't know anything about. Here is a tool which compares the same position across multiple tracks.
OK, so suppose I want a track of named quinoa genes over its genome?
No one appears to have a write-up for going directly UniProt to jBrowse. This got me concerned.
Then I discover Enseml Plants has a browser with quinoa genes marked on it:
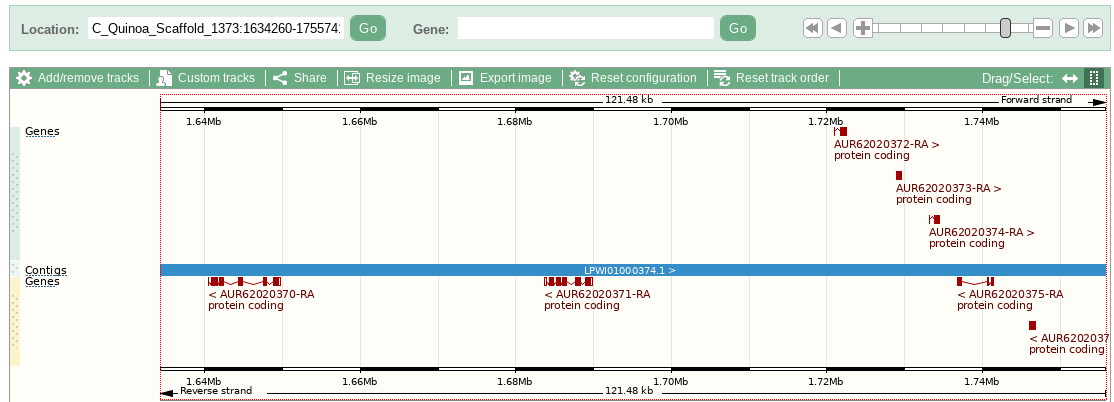
So where did these come from..? Do any have more detail?
On UniProt the quinoa proteins have a Genomic Coordinates tab, giving Assembly Name "ASM168347v1" and Genomic location "3,770,659–3,784,205". What's happening:
- ASM168347v1 is a genome uploaded by KAUST (Saudi Arabia) in 2017, and it's the NBCI reference genome for the species. This seems to be the source of the thousands of unreviewed quinoa proteins in UniProt and annotations on Ensembl.
- The locations are a specific index within this read, which was not broken into chromosomes.
- On this page , I can download annotations to the genome in a GFF file. This annotation is useful to me because it identified coding portions of the DNA, has IDs connectable to UniProt, and has some initial notes on similarity to other reference plants. But if I load a newer quinoa genome into jBrowse with chromosomes, it cannot smoothly match the locations.
- Maybe there's a way I could convert the protein amino acids -> DNA sequences and relocate them on a newer genome ?
- The few kaniwa proteins on UniProt have the Genomic Coordinates section grayed out, possibly because they belong in chloroplasts.
The jBrowse website invites users to set up office hours, so I booked one.
Meanwhile! I was considering contacting quinoa / kañiwa experts about the mystery of huazontles. Then I find this paper published days ago on May 29th - the genome is read and the researchers are convinced that huazontles shares a common, polyploid ancestor and quinoa split off on a later southern migration. The genome is in many pieces (contigs) and not chromosomes, but that's fine. I can download the genome here (this was uploaded in December but I didn't find it in searches, maybe because nuttaliae is in the subspecies field).
NCBI also has 129 named genes. After review, these were easier to identify because they're from the chloroplasts and match other plants.
The next morning, I download the genome and these genes, to prove to the jBrowse people that I am not totally lost. I wrote a script to generate a GFF file, placing these over the right sequence locations with the right contig ID + numeric offset.
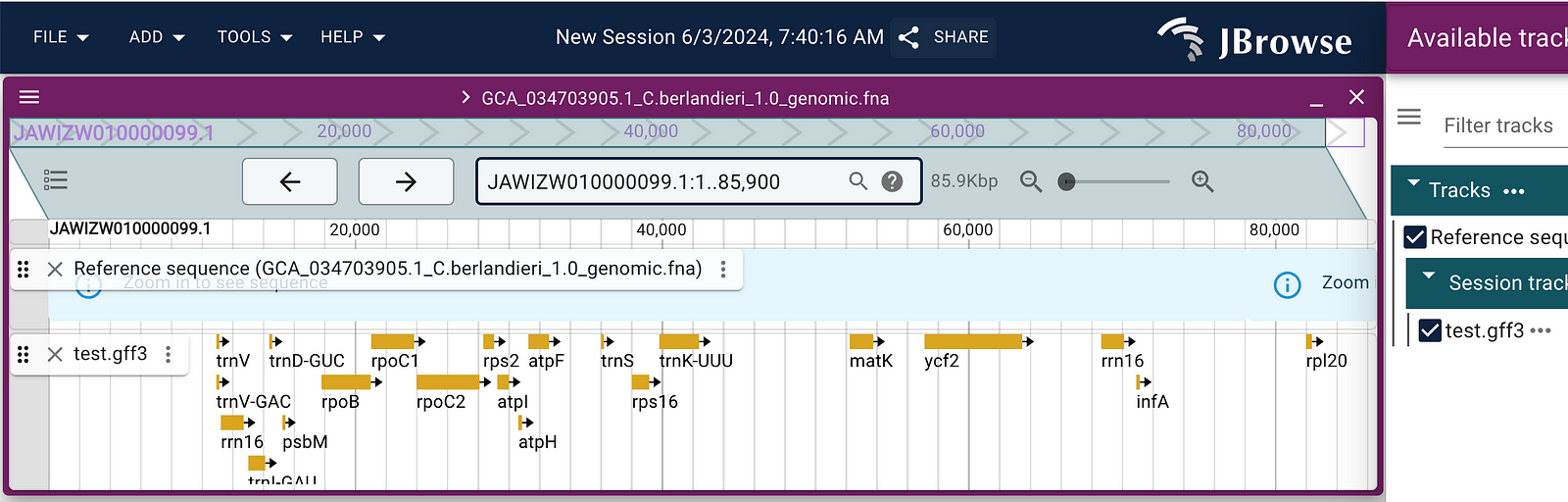
By default I had all genes/annotations appear to go left-to-right. But when we zoom in, we see these green and red colored lines:
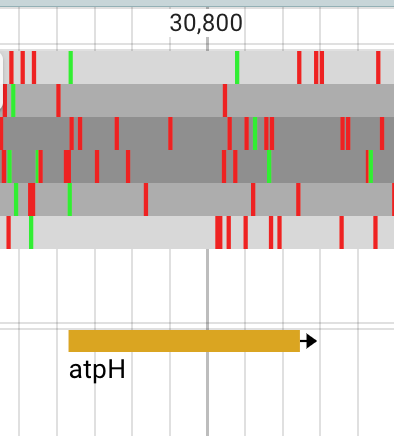
These represent start and stop codons for 3 different frames x 2 directions of reading. atpH could be left-to-right based on the green and red marks at the ends.
Handcoding which contigs to search for in which gene sequences, then formatting out a GFF, is not a great strategy TBH. There's got to be tools which do this automatically. Or maybe I run the tool to mark the start and end of all plant protein-coding genes, and then try to match them to known genes/proteins.
Finally I did the jBrowse call today. This was better received than the CSV Conf talk and cleared up some of my confusion. Some of my notes plus research here:
- jBrowse is usually run internally.
- My idea of a wiki / participatory site is something that the developers discuss. But for something like quinoa, suppose there is a potential audience of 100 and I need 10+ to participate.
- Gene Wiki (2022 retrospective) recruited specialists to write articles about genes.
- WikiGenes was a project connecting directly into Wikipedia / WikiData, but interest tapered off 10 years ago
- OntoGPT has a project "Transforming unstructured biomedical texts with large language models"; I should watch the video.
- Gene prediction tools will be helpful for adding genes or for placing known genes. I had seen some of these plus jBrowse recommended a deep learning repo.
- My GFF file should add rows defining the sequence, which will make it easier to place in the colorful view.
I gave my feedback about lacking a "zoom to" feature for custom tracks, lacking a way to favorite/star/direct link a specific contig (there are 150+ contigs in huazontles and only 2 where I have tracks), and an error reporting issue.