Georeactor Blog
RSS FeedQuantizing Evo?
I've been attempting to do more DNA model experiments by applying int8 quantization to Evo. The accuracy can be compared to the original Evo model based on whether the log-likelihood scores are similar for a set of DNA sequences in FASTA format.
The Evo repo comes with a script to calculate log-likelihood on three very short sequences. The values from last month's quantized model and the full-size model didn't match, and actually weren't in agreement of least to greatest. I had trouble finding my past notebooks and knowing whether I had used the "1.1_fix" Evo model as a starting point, so I redid int8 quantization on the 1.1 model.
Given these are very short sequences, I used a script to extract 150-base-pair excerpts, 5 samples x 100 contigs from the giant kelp genome. This is a eukaryote genome, but as an algae it's a little less weird.
Looking at a scatter plot, the scores do not look correlated:
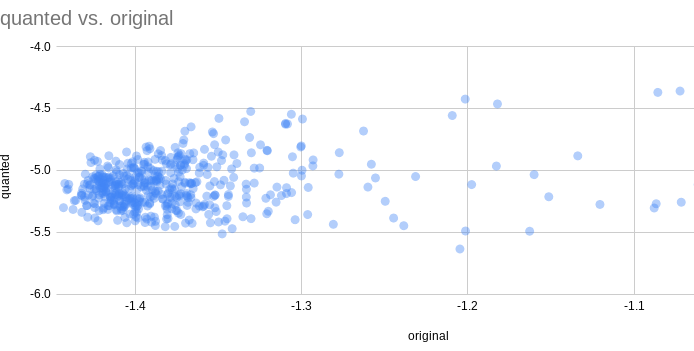
One of the possible issues was a line in the Evo repo, model.to_bfloat16_except_poles_residues(), but without the conversion there is a type mismatch.
Other issues might be code or config files being pulled from the original Evo, without the quantization settings.
So moving forward in this, I'll probably work with Evo on an L4 GPU instead of trying to make claims with the quantized version.
Notebook: https://colab.research.google.com/drive/1J1HTxj9UqiaU-D3bwqyloKRA5rtyy75h?usp=sharing